Plants can recognize pathogens and respond to them. Roger Innes, Distinguished Professor in the Indiana University College of Arts and Sciences Department of Biology, and members of his lab are investigating the molecular and cellular basis of disease resistance in plants. They want to understand how plants detect pathogens and how detection then activates immune responses.
Innes lab researchers and colleagues in the lab of Blake Meyers, a member of the Donald Danforth Plant Science Center in St. Louis and Professor of Plant Sciences at University of Missouri-Columbia, recently discovered that the leaves of plants accumulate RNA on their surfaces and in the spaces between cells.
RNA is usually thought of as a molecule that can direct cells to synthesize specific proteins. An example that has frequented the news in the last two years is the messenger RNA (mRNA) in COVID-19 vaccines that directs human cells to make SARS-CoV2 spike protein. Some RNAs, however, serve other functions.
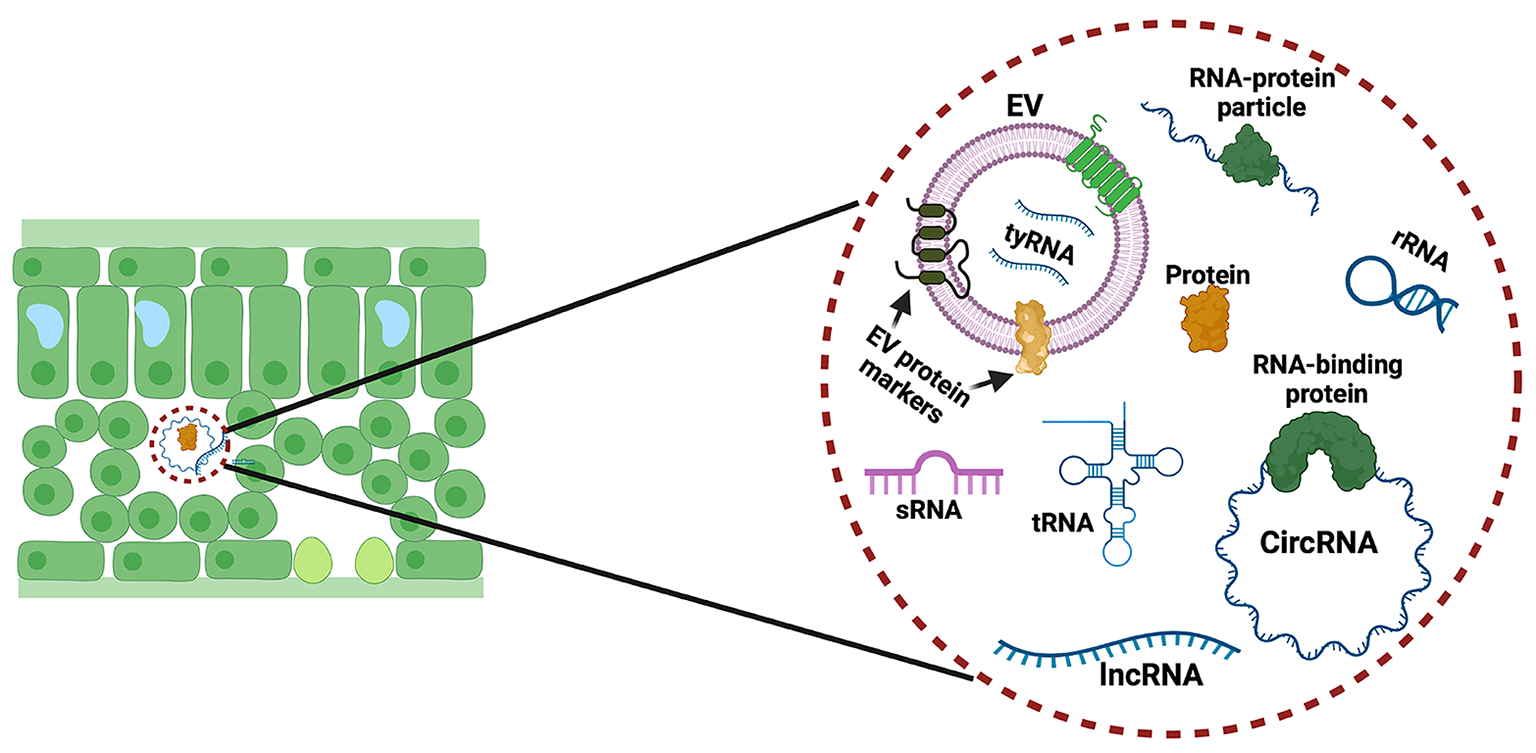
To prevent infection by disease-causing microbes, plants secrete diverse antimicrobial compounds into their extracellular spaces. Included among these compounds are small RNA molecules 21 to 24 nucleotides in length (sRNAs) that can be taken up by microscopic organisms such as bacteria. The sRNAs are thought to cause the destruction of mRNAs with complementary sequences. The processes by which sRNAs are secreted, how they are protected from degradation, and how they are taken up by pathogenic microbes are all poorly understood.
Although extracellular sRNAs had been shown to co-purify with extracellular vesicles (EVs), it had not been clearly established whether sRNAs were located inside or outside EVs, or whether plants secreted RNAs longer than 24 nucleotides.
Researchers in the Innes lab isolated EVs from the extracellular spaces in the leaves of Arabidopsis thaliana (a species of mustard plant) and then treated the preparations with enzymes that degrade proteins and then with different enzymes that degrade RNA. The RNA remaining after these treatments was then analyzed by RNA sequencing performed by the IU Center for Genomics and Bioinformatics and the Meyers laboratory. These analyses revealed that sRNAs are associated with protein complexes located outside EVs. Significantly, they found that Arabidopsis secretes both sRNAs and much longer RNAs, from 30 to over 500 nucleotides in length. The longer RNAs do not code for proteins, and many have a circular structure. Notably, both the long non-coding RNA and the small RNAs were found to be highly enriched in a post-transcriptional modification known as N6methyladenine (m6A). The researchers speculate that this modification might be required for secretion of RNA.



 The College of Arts
The College of Arts