- Postdoctoral Fellow, University of California, Berkeley, 1988-1991
- Ph.D., University of Colorado, 1988

Roger Innes
Distinguished Professor, Biology
(he/him/his)

Distinguished Professor, Biology
(he/him/his)
Myers Hall 302
812-855-2852
Member, National Academy of Sciences
Fellow, American Academy of Microbiology
Fellow, American Association for the Advancement of Science
Career Achievement Award, International Society of Molecular Plant-Microbe Interactions
Recipient, Indiana University Bicentennial Medal
Our primary interest is in understanding the molecular and cellular basis of disease resistance in plants. Plants are able to specifically recognize pathogens and actively respond. We are investigating how this specific recognition is accomplished and how recognition is translated into a resistant response. To address these questions we take a molecular genetic approach. We use the small mustard Arabidopsis thaliana as our standard host plant, and both fungal (powdery mildew) and bacterial pathogens ( Pseudomonas syringae) as our standard pathogens. Recognition of specific P. syringae strains by Arabidopsis is mediated by specific disease resistance (R) genes of Arabidopsis. These R genes encode intracellular receptors that detect a signal produced directly or indirectly by bacterial proteins that are injected into the plant cell. Our laboratory has made significant contributions to our understanding of HOW R proteins mediate pathogen recognition. These insights are now leading to new approaches for engineering disease resistance in plants. For example, we have shown that the R protein RPS5 is activated by proteolytic cleavage of a second host protein PBS1 by proteases secreted by P. syringae. Armed with this knowledge, we are now engineering this system to recognize new pathogens by modifying the protease cleavage site within PBS1 so that it becomes cleavable by proteases from other pathogens, such as viruses. In this manner, we believe we can engineer resistance to a diverse array of pathogens, once we know the proteases employed by these pathogens to cause disease. We are currently applying this discovery to engineer novel disease resistance traits in soybean.
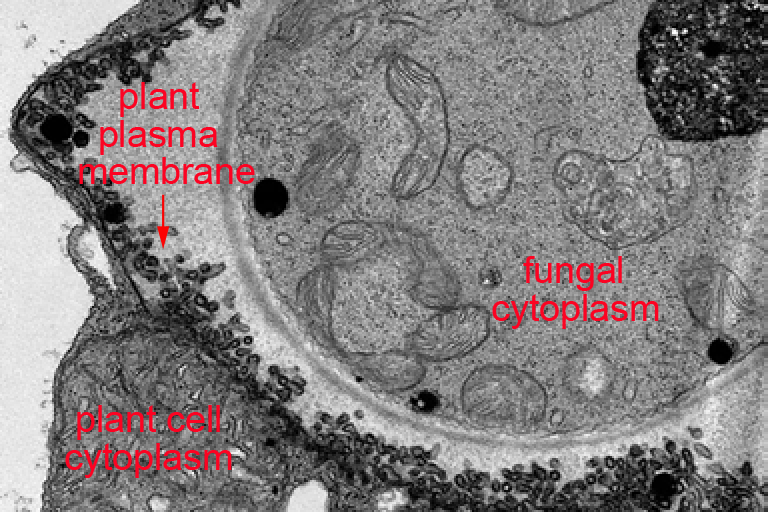
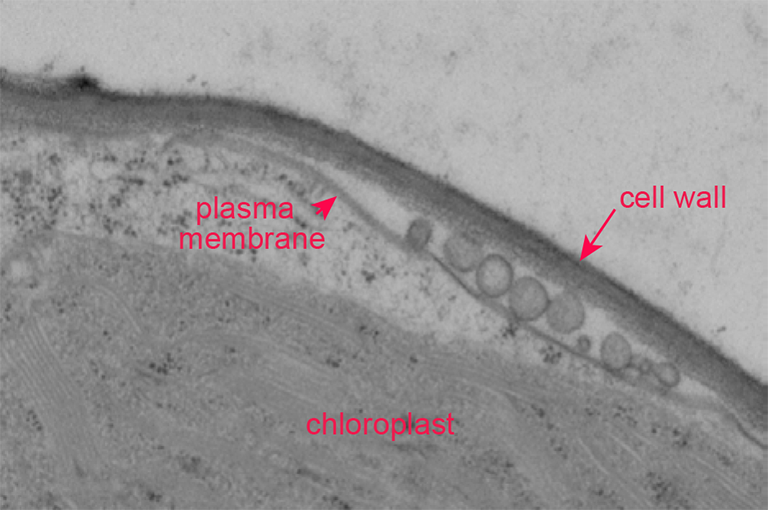
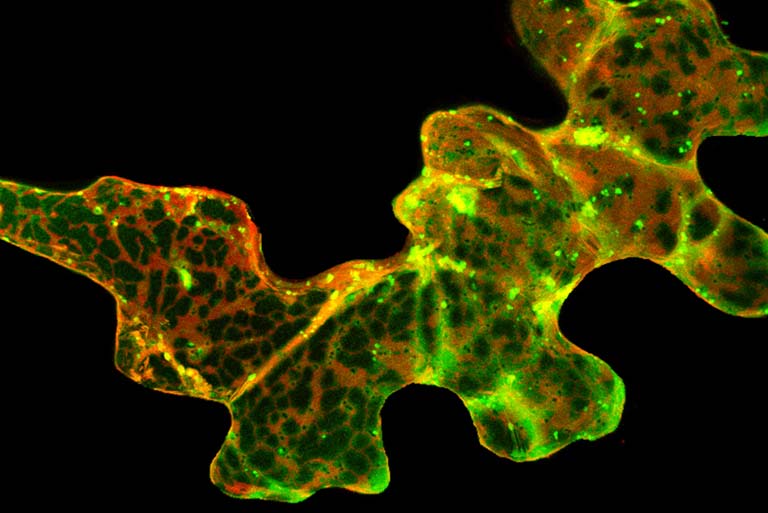
In a second project, we have been examining endomembrane trafficking in plant cells in the context of active defense responses. When a plant leaf is colonized by a pathogen, it dramatically increases secretion of antimicrobial compounds. This secretion process is complex and appears to involve non-canonical secretory pathways. Based on electron microscopy analyses, this process includes secretion of vesicles known as exosomes, which are lipid-bilayer spheres ranging in size between 50 and 200 nm. The contents of these vesicles and their function in immunity has not been reported. We have recently determined that they are enriched in defense proteins and carry microRNAs. This latter finding is quite exciting as it suggests plants may employ exosomes for intercellular signaling, and perhaps interkingdom signaling. We hypothesize that exosomes may mediate transfer of silencing RNAs between plants and pathogens, and are now actively testing this hypothesis.
Eukaryotic Cell Biology, Cytoskeleton, and Signaling
Microbial Interactions and Pathogenesis
Plant Molecular Biology