Editor's note: Sofía Casasa, a postdoctoral fellow in the laboratory of Assistant Professor Erik Ragsdale in the Indiana University College of Arts and Sciences Department of Biology, writes about the research she performed as a graduate student in Professor Armin Moczek's laboratory in the department. Collaborating on the project was Eduardo Zattara, who had been a postdoctoral researcher in the Moczek lab and is now an adjunct researcher at INIBIOMA-CONICET in Argentina as well as a research associate at Indiana University and at Smithsonian Institution. Their landmark research focuses on plasticity—the adaptability of an organism to changes in its environment or differences between its various habitats—and was published in Nature Ecology & Evolution.
How do organisms interact with their environments? Research on phenotypic plasticity focuses on how organisms with the same genotype can adjust their phenotypes in response to different environmental conditions. Plasticity can take many forms and can occur at any stage during development, but those changes that occur early in development are commonly irreversible and it is those changes that fall under the rubric of developmental plasticity.
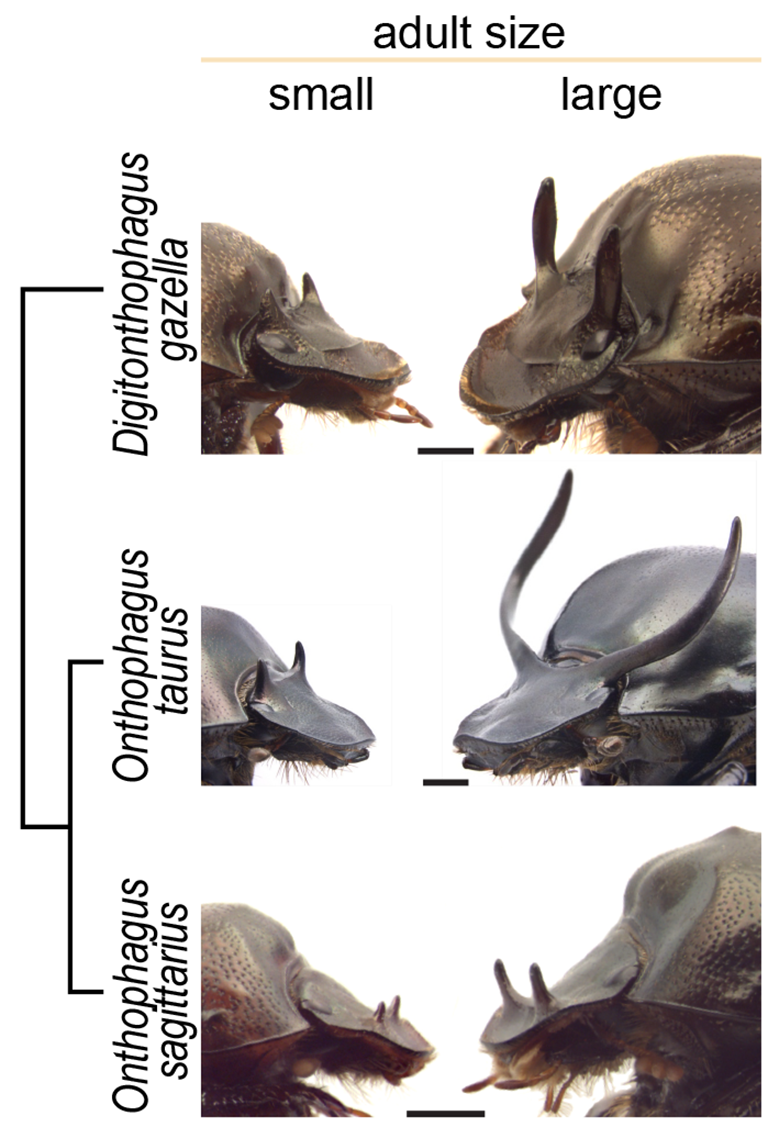
An extreme case of developmental plasticity are polyphenisms, where instead of showing a gradual response to a particular environmental factor, individuals develop into distinctly different, alternative phenotypes. One of the most well-known examples of polyphenism are castes in social insects (bees, ants, etc.), in which nutritional conditions determine whether females with the same genotype develop into workers or queens, with no intermediates.
Yet, what are the mechanisms that specify development of one or the other phenotype? And, how does plasticity itself evolve? Are the genetic mechanisms underlying plasticity responsible for the different degrees of plasticity seen across species?
I have been tackling these questions for the past several years. Our recently published article in Nature Ecology & Evolution was the last chapter of my Ph.D. dissertation, and tried to answer some of these questions from a comparative transcriptomic approach. My advisor, Armin Moczek, and I decided to collaborate with Eduardo Zattara, a former postdoctoral researcher in the lab who has ample experience in analyzing transcriptomes. Despite having returned to Argentina, Eduardo was the perfect mentor to remotely guide me through all the bioinformatic tools needed for this project.



 The College of Arts
The College of Arts