- Postdoctoral Fellow, Harvard Medical School, 2009-2017
- Postdoctoral Fellow, University of Oxford, 2007-2009
- D.Phil., University of Oxford, UK, 2007

Xindan Wang
Associate Professor, Biology
(she/her/hers)

Associate Professor, Biology
(she/her/hers)
Biology Bldg. 225
Wang Lab website
Indiana University Outstanding Junior Faculty Award, 2023
Indiana University Trustee’s Teaching Award, 2020
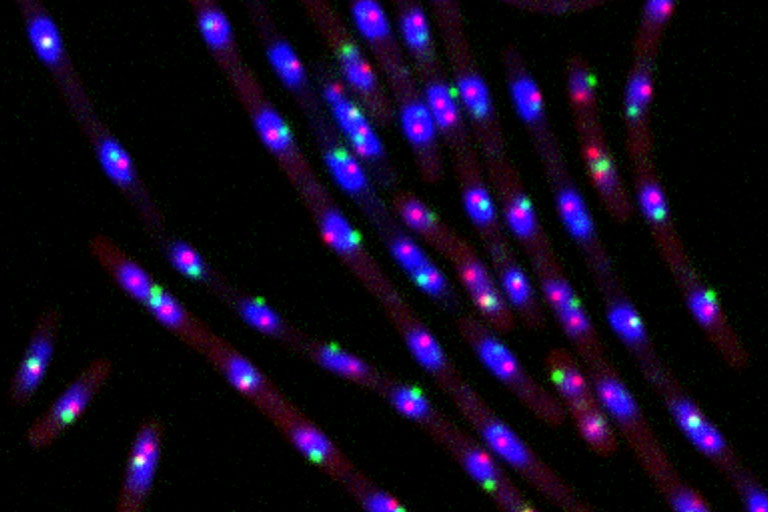
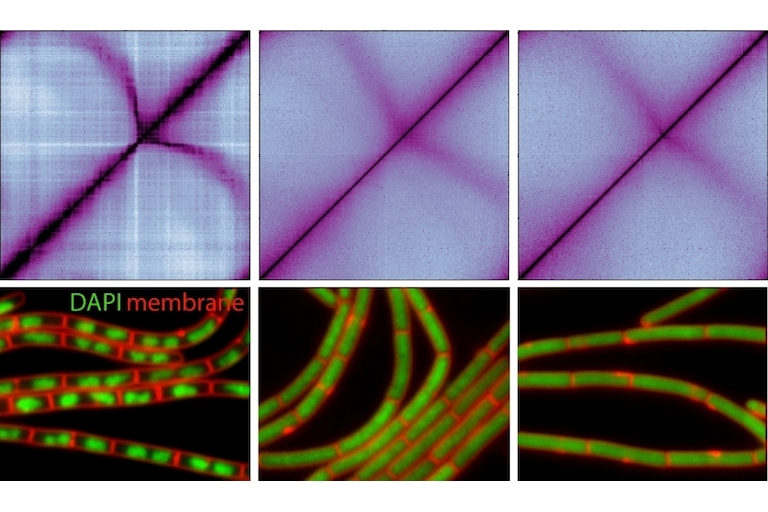
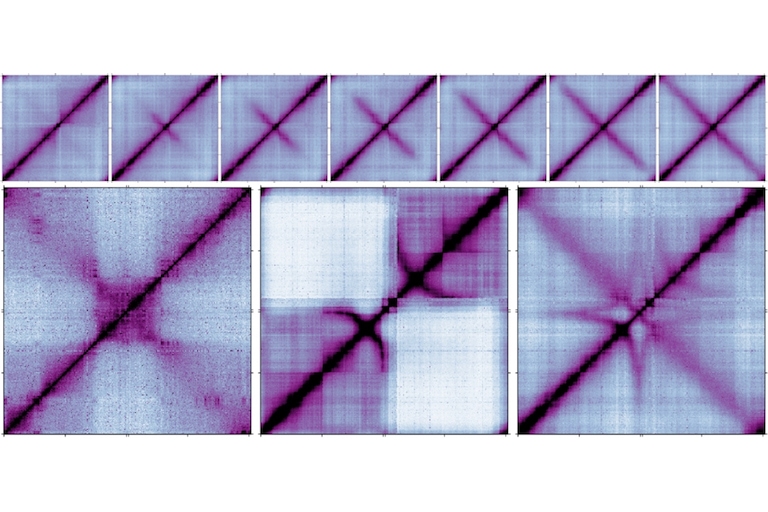
Our lab is interested in chromosome dynamics in bacteria. Using Bacillus subtilis as a model organism, we combine genetic, molecular, cytological, and biochemical approaches to study how the chromosome is organized and segregated during the cell cycle. Our research primarily focuses on a set of highly conserved factors in chromosome biology, including the parABS partitioning system and the SMC condensin complex. Taking the many strengths of studying bacteria, our goal is to uncover general principles of chromosome organization and segregation in living organisms. Specific research directions include:
Chromatin, Chromosomes, and Genome Integrity
Genomics and Bioinformatics
Microbial Cell Biology and Environmental Responses