Lab
Simon Hall 011
812-855-5209
Awards
IU Distinguished Faculty Research Lecturer, 2023
Fellow, American Association for the Advancement of Science, 2018
Associate Editor, PLoS Pathogen, 2017
Board of Editors, mBio, 2016
Division T Chair, American Society for Microbiology, 2012
Fellow, American Academy of Microbiology, 2012
Research
Rotaviruses are a large genetically diverse and rapidly evolving population of icosahedral viruses with genomes consisting of segmented double-stranded RNA. These viruses are major causes of severe diarrheal disease in the young of many animal species, including humans, pets, livestock, and birds. In infants and young children, rotavirus infections account for approximately 200,000 deaths each year, most occurring in developing countries of Africa and Asia. Several developed countries, including the US, have introduced rotavirus vaccines into their childhood immunization programs; these vaccines that have been remarkably effective in preventing severe diarrheal disease. However, in developing countries, these same vaccines have been significantly less effective for reasons that remain unclear. Our laboratory studies rotavirus molecular biology and pathogenesis, in hopes of providing fundamental knowledge useful for advancing new ways of treating and preventing rotavirus disease.
Virus-host interactions. Activation of innate immune system creates an environment in mammalian cells that is hostile to virus replication. Activation of this system depends on the expression of interferon (IFN), a signaling protein necessary for the production of numerous antiviral IFN-stimulated gene (ISGs) products. In rotavirus-infected cells, IFN signaling is inhibited by the action of NSP1, a major virulence determinant of the virus that induces the degradation of host proteins essential for IFN expression. NSP1 mediates degradation by hijacking cellular E3 ubiquitin ligases, causing this machinery to inappropriately tag host proteins for degradation by proteasomes. A major interest of our laboratory is to understand the mechanisms used by NSP1 to seize control of E3 ligases and to recognize cellular degradation targets.
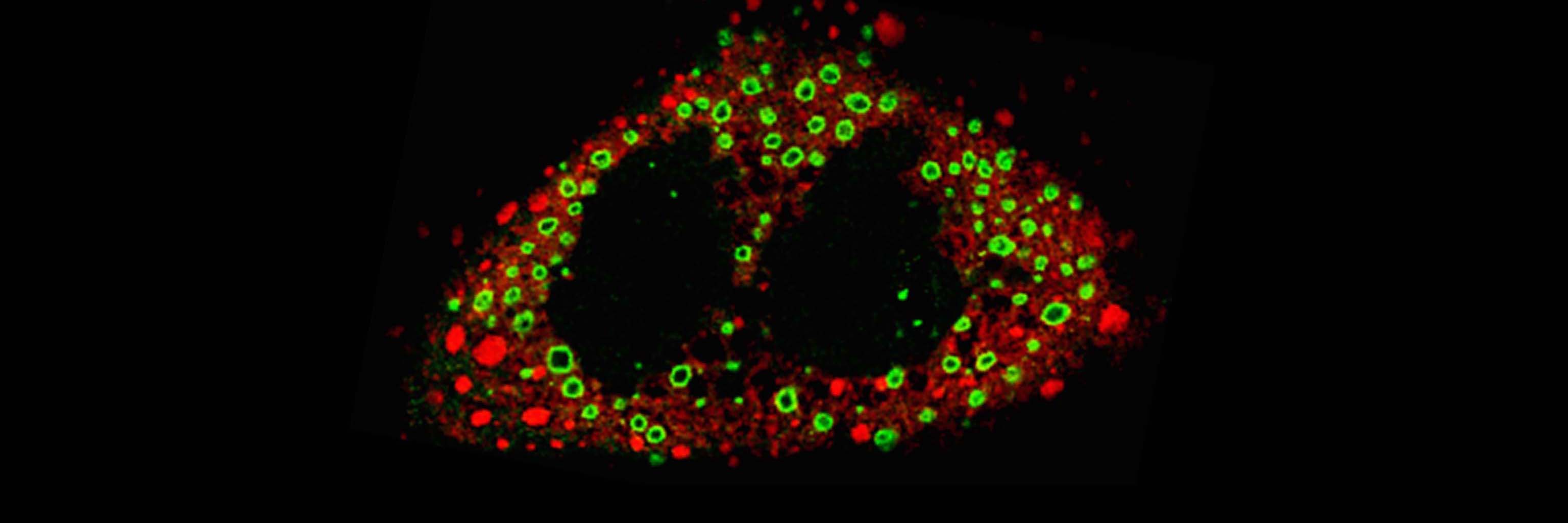
Viral RNA packaging and replication. The cytoplasm of rotavirus-infected cells contains many large electron-dense inclusion bodies, termed viroplasms. These structures serve as viral factories, where the segmented double-stranded RNA genome of the virus is packaged into progeny cores during synthesis by the viral RNA polymerase. The coordinated nature of rotavirus genome replication and packaging results from allosteric regulation of the viral RNA polymerase by the core capsid protein. The polymerase is only active when sufficient core capsid protein is available to package newly made double-stranded RNA into progeny cores, thus assuring that uncoated dsRNA—an inducer of IFN expression—is not produced in infected cells. In our laboratory, we pursue studies designed to understand the organization and activities of viroplasms, the function of the polymerase and other proteins that contribute to genome replication and progeny assembly, and the structure of rotavirus replication intermediates.
Re-engineering the rotavirus genome. The rotavirus genome consists of 11 segments of dsRNA and codes for a total of 12 proteins. With the recent development of a complete plasmid-based reverse genetics system for rotavirus, re-engineering of the genome is now possible. In our laboratory, we are modifying the rotavirus genome such that it expresses a 13th protein. As a result, we can engineer rotavirus strains that can produce capsid-protein components of other enteric viruses, for example, norovirus. Through this route, we hope to create a next generation rotavirus-based combination vaccine that induce protective immune responses against the two most significant causes of childhood diarrhea, rotavirus and norovirus.
Research Area
Virology

 The College of Arts
The College of Arts